Pythiomics API
Seamless Queries. Seamless Computation.
Get programmatic access to Pythiomics — our trusted, well-curated multi-omics database — directly from your native environment.

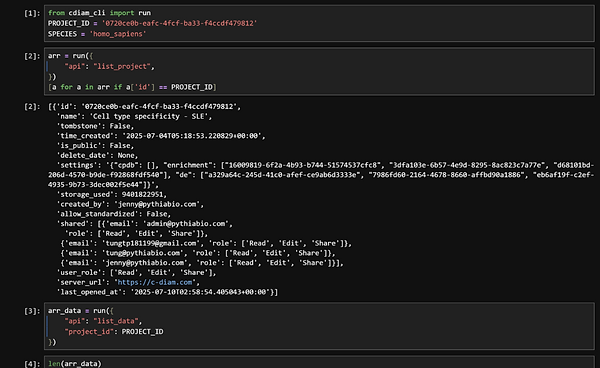
Access, Query, and Compute with Confidence
The Pythiomics API is a software package that provides programmatic access to Pythiomics, our well-curated and standardized multi-omics database of more than 10,000 datasets. It enables retrieving and quering the database directly from native bioinformatics environments such as Jupyter Notebook and RStudio - and receiving results in various formats, including text-based matrix files, XML, or JSON.
Programmatically Explore
10,000+ Multi-omics Datasets
Explore a standardized and reliable data source of comprehensive multi-omics data meticulously curated, harmonized and QC'ed.
Bulk RNA
Visium HD & Xenium
Coming soon
Single-cell RNA
Metabolomics
Coming soon
Proteomics
ATAC-Seq & CITE-Seq
Coming soon
Visium Spatial
CHIP-Seq
Coming soon
CosMx
Mutation & GWAS
Coming soon
Tailor-made for Computational Biologists and Bioinformaticians
Whether you're building custom workflows or running large-scale analyses, Pythiomics API puts trusted omics data at your fingertips — ready to explore, compute, and publish.
Flexible Access. Flexible Analytics.





